
So this is how you can really zoom in and look at just the If you were just looking in your DNA pulled out of your cell, that would be a needle in a haystack. So you want to make copies, or as they say amplify it, so that you could run it in gels and stuff and see how all of those molecules, how big they are or something like that. It's hard to identify just one fragment of that gene. To a particular condition, all sorts of really To see if you have a gene that would predispose you In medical diagnostics, so this could actually be your DNA that was being checked Used a lot in forensics, it's also used a lot Where else would you have to do PCR? - PCR is And you might start with a very small sample of DNA. Sticking one fragment into one plasmid, you're doing that with many, so you need a lot of fragments of DNA. We talked about cloning, and we're talking about sticking a fragment of DNA inside of a plasmid, it's not like you're just To be making lots of copies so that you can clone it into a plasmid, and then do some otherĮxperiments with it, that's a big use. Why would you need to make a lot of copies of a particular fragment of DNA? - So you might want Makes you a lot of copies of a particular fragment ofĭNA that you're interested in. And so whatĭoes PCR in particular do? - PCR basically Why have you done PCR? - PCR was kind of the mainstay of my graduate project, where I built all sorts of different recombinant DNA molecules, and used them to learn I'm here with Emily, our biology content fellow, to talk about PCR, or Polymerase Chain Reaction, which you've actually done a lot of. (Replication follows a similar pattern, though of course using different enzymes.) This bubble is moved along the DNA by the RNA polymerase complex as it transcribes the DNA. These activities open up what is known as a "transcription bubble" - a short (around 13 nt) region where the two strands of the DNA double helix are separated from each other. In eukaryotes this complex also temporarily "unpacks" the DNA from chromatin, so this is quite a complex process. The process seems to involve the RNA polymerase complex unwinding and forcing the strands apart. However, you aren't completely wrong - promoters contain A:T-rich sequences that are easier to separate ("melt") due to A:T base pairs having a weaker interaction compared with G:C base pairs. A cell would have to raise the temperature well over 90☌ to have a significant effect on chromosomal DNA, so that probably would kill the cell assuming it had enough energy. Interesting idea, but the separation happens without any significant heating. §Note that the strength of interaction between G:C pairs is stronger that for A:T pairs, so the "GC content" also matters. Primers in that length range typically bind best in the 50-65☌ range. Primers around 20-25 nt long generally show good specificity and are relatively inexpensive, so that is what typically gets used. Primers can be made that bind at the extension temperature (72☌), but longer primers are more difficult to make and thus more expensive. This reduces the chance that the primer will bind to the wrong sequence. This means that interactions between primers and a sequence they don't perfectly match will denature at a lower temperature.Ĭonsequently, primers are designed so that they can only efficiently bind to a perfectly matched sequence at relatively high temperature - e.g. The strength of a primer-target interaction affects how resistant it is to denaturation (i.e.
As you line up more correct pairs the interaction between the primer and its target sequence gets stronger, so longer primers tend to bind more strongly than shorter primers§. You can think of each base pair as being like the interaction between one hook and one loop in velcro. Primers bind to longer nucleic acids by making base pairs.
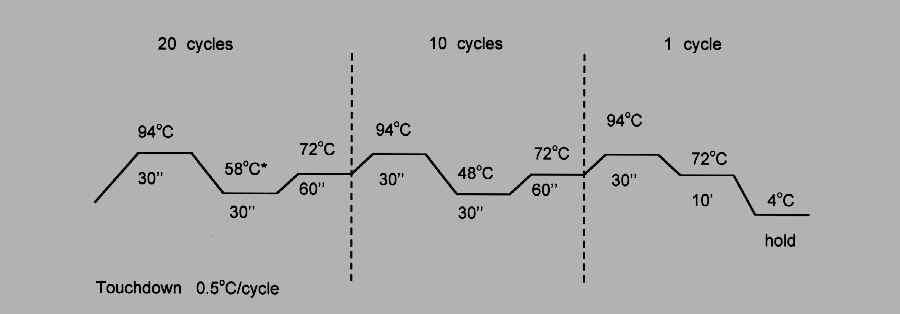
The first annealing between primer and DNA occurs at the initial temperature, the second after the temperature has been lowered by Δ T.


 0 kommentar(er)
0 kommentar(er)
